Hi!
As a zoologist, I often get asked the classic question: “Soooo, what’s your favorite animal?”
And honestly, I think that question should be banned. It’s like asking parents whether they have a favourite. Except, as a zoologist, I don’t just have a couple of options. I have thousands to choose from.
So, no, I don’t really have a favourite. But every now and then, I come across a creature so bizarre and fascinating that I can’t help but be obsessed for a while.
Right now, I’m in my Colugo Era. Seriously, just look at them!
Fun fact: Colugos are sometimes called flying lemurs, but that’s pretty misleading. For one, they don’t actually fly — they glide. And two, evolutionarily speaking, they’re not lemurs at all. Despite looking a lot like flying squirrels or sugar gliders, their closest relatives are primates. Basically, they are just little pranksters of the animal kingdom.
Okay, back to the topic!
We’ve learned how to filter and search for specific letters and numbers, but what if you need to figure out what sits in a very specific spot in your dataset? It’s kind of like playing Battleship.
What’s in F2?
The only difference is that we don’t use letters for columns in this case. Instead, you’d say: “What do you have in row 6, column 2?” And if we were to translate this into R language, it would look something like this (grab the dataset here):"
### Libraries ###
library(readr)
### Loading data ###
blue_tits <- read_csv("paste_the_path_to_the file_here")
### Searching for the specific value ###
blue_tits[6,2]You could also ask: “What is in row no. 6 in the Laying_day column?”
blue_tits["Laying_day"]You could also ask many other questions so please play by typing all these commands into the console:
blue_tits[1,] # first row all columns
blue_tits[,1] # all rows of the first column
blue_tits[5,4] # 5th row, 4th column
blue_tits[4:11,2] # a range of elements (4 and 11 inclusive) in the 2nd column
blue_tits[5:11,"Laying_day"] # a range of elements (4 and 11 inclusive) in the 2nd column, just differentlyAnd okay, maybe you are wondering whether it is worth the hype, but yes, it will be very useful in your R journey! For example, when you want to remove a row or column or multiple rows and columns, you can type:
blue_tits[,-2]
blue_tits[-1,]
blue_tits[-(2:100),2]The caveat here is that you cannot remove a single element without destroying the dimensions of the data frame (and we don’t want our data frames to look like slices of Swiss cheese). However, you can assign it the “NA” value.
blue_tits[1, 2] <- NAOkay, now we are gonna try something else. What about unpacking a package that we took from a bigger package? Let’s try:
output <- cor.test(blue_tits$Laying_day, blue_tits$Hatching_day, method = "pearson")
# Let's see whether hatching dates correlate with laying dates (they should!)
# Please, let's forget about the assumptions of the Pearson's correlation test for now. Just not in your stats class.Now type “output” in the console.
What do you see? Does it look like:
Okay, how can we display only the p-value? Any ideas?
Let’s experiment with “output[x]”, where x is the number you suggest.
And what about:
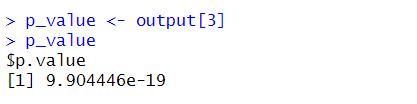
p_value <- output[3]You should get:
And now, how can we get just the value, 9.904446e-19, without the “$p.value” part?
The ”[ ]” brackets tell R to “unpack something” and our value of interest sits in a box that sits in a bigger box.
We can see that our value is the only one here, so it sits in the first row of the first column with the “$p.value” attached to it like a label. To get rid of the label (the fancy name of this action is to “drop name attribute”), type:
p_value[[1]]Hurray! We got it! This is cool news, because now if you ever had a crazy desire to perform any arithmetic operation on our p-value, you can do it. But don’t just believe me (who knows maybe I am a prankster, like a colugo), try:
p_value[1] + 3
#VS
p_value[[1]] + 3One last thing to keep in mind before you go:
In R, counting starts at 1, but in other languages like Python 🐍, it starts at 0!
See you soon,
Aga
PS: Here is the survey in which you can tell me what R topic you find particularly confusing and why you want to learn it so that we can shape this space together!